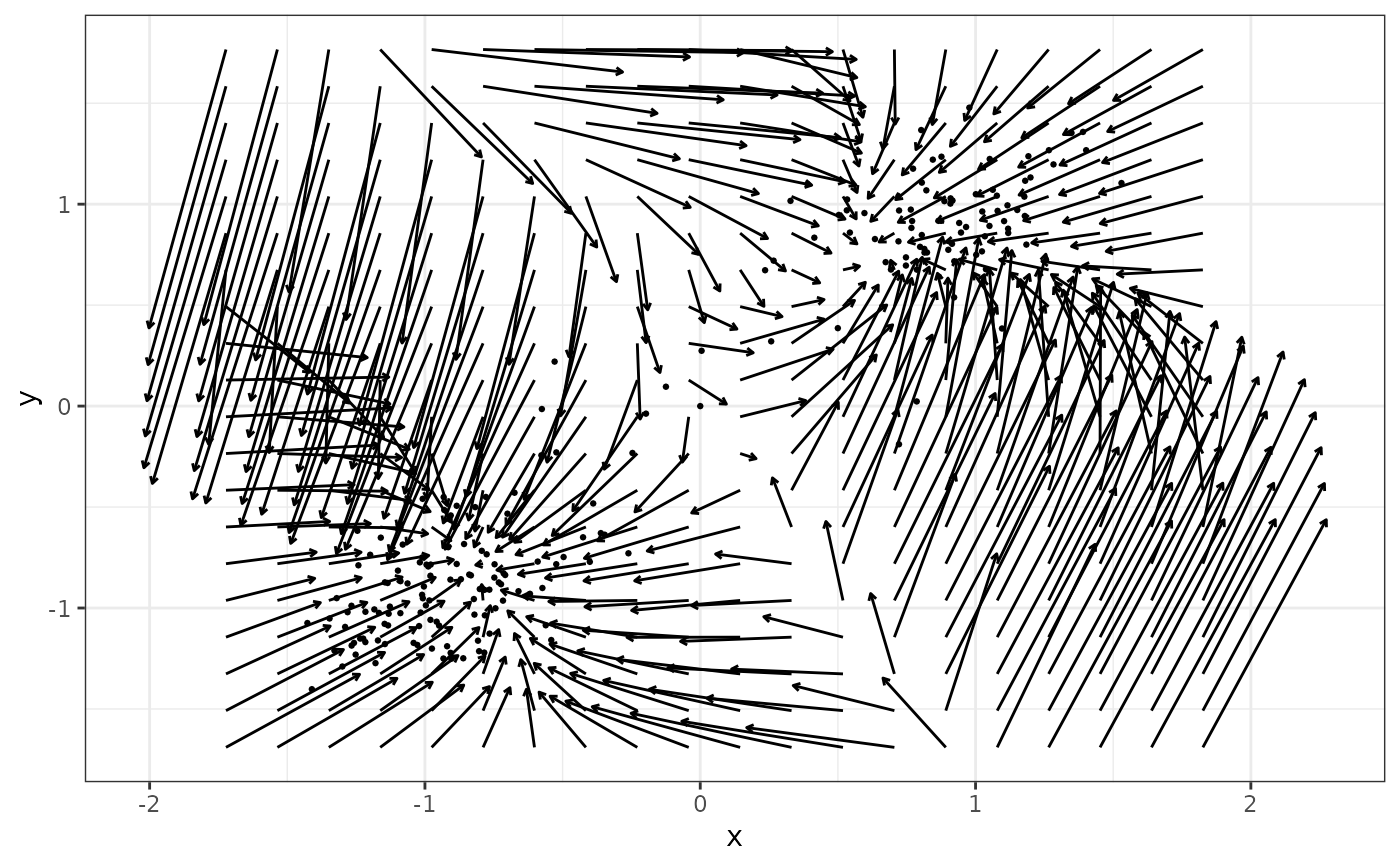
Estimate a 2D vector field from intensive longitudinal data. Two methods can be used: Multivariate Vector Field Kernel Estimator (MVKE, using MVKE()), or Sparse Vector Field Consensus (SparseVFC, using SparseVFC::SparseVFC()). Note that the input data are automatically normalized before being sent to the estimation engines to make sure the default parameter settings are close to the optimal. Therefore, you do not need to scale up or down the parameters of MVKE() or SparseVFC::SparseVFC(). We suggest the MVKE method to be used for psychological data because it has more realistic assumptions and produces more reasonable output.
Usage
fit_2d_vf(
data,
x,
y,
lims,
n = 20,
vector_position = "start",
na_action = "omit_data_points",
method = c("MVKE", "VFC"),
...
)Arguments
- data
The data set used for estimating the vector field. Should be a data frame or a matrix.
- x, y
Characters to indicate the name of the two variables.
- lims
The limits of the range for the vector field estimation as
c(<xl>, <xu>, <yl>, <yu>). If missing, the range of the data extended by 10% for both sides will be used.- n
The number of equally spaced points in each axis, at which the vectors are to be estimated.
- vector_position
One of "start", "middle", or "end", representing the position of the vectors. If "start", for example, the starting point of a vector is regarded as the position of the vector.
- na_action
One of "omit_data_points" or "omit_vectors". If using "omit_data_points", then only the
NApoints are omitted, and the points before and after anNAwill form a vector. If using "omit_vectors", then the vectors will be omitted if either of its points isNA.- method
One of "MVKE" or "VFC".
- ...
Other parameters to be passed to
MVKE()orSparseVFC::SparseVFC().
Examples
# generate data
single_output_grad <- simlandr::sim_fun_grad(length = 200, seed = 1614)
# fit the vector field
v2 <- fit_2d_vf(single_output_grad, x = "x", y = "y", method = "MVKE")
plot(v2)